The University of Liège wishes to use cookies or trackers to store and access your personal data, to perform audience measurement. Some cookies are necessary for the website to function. Cookie policy.

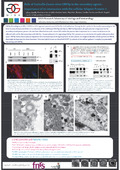  | Lebrun, M., riva, L., lambert, J., Rambout, X., Thiry, M., blondeau, C., Twizere, J.-C., Dequiedt, F., & Sadzot-Delvaux, C. (20 May 2016). Role of Varicella Zoster virus ORF9p in the secondary egress : importance of its interaction with the cellular Adaptin Protein-1 [Poster presentation]. 2015 Colorado Alphaherpesvirus Latency Society Symposium, Vail, United States - Colorado. |
  | Rambout, X., Detiffe, C., Bruyr, J., Mariavelle, E., Cherkaoui, M., Brohee, S., Demoitie, P., Lebrun, M., Soin, R., Lesage, B., Guedri, K., Beullens, M., Bollen, M., Farazi, T. A., Kettmann, R., Struman, I., Hill, D. E., Vidal, M., Kruys, V., ... Dequiedt, F. (2016). The transcription factor ERG recruits CCR4-NOT to control mRNA decay and mitotic progression. Nature Structural and Molecular Biology. doi:10.1038/nsmb.3243  Peer Reviewed verified by ORBi Peer Reviewed verified by ORBi |
Lebrun, M., riva, L., Rambout, X., Thiry, M., Blondeau, C., Twizere, J.-C., Dequiedt, F., & Sadzot-Delvaux, C. (26 July 2015). Identification of VZV ORF9p potential cellular partners that could be important for the viral egress [Poster presentation]. 40th international herpesvirus workshop, boise, United States - Idaho. |
  | Detiffe, C., Dequiedt, F., & Rambout, X. (Other coll.). (June 2015). The class IIa HDACs prevent degradation of RBFOX2 by Chaperone-Mediated Autophagy [Poster presentation]. Autophagy, Breckenridge, United States - Colorado. |
  | Rolland, T.* , Tasan, M.* , Charloteaux, B.* , Pevzner, S. J.* , Zhong, Q.* , Sahni, N.* , Yi, S.* , Lemmens, I., Fontanillo, C., Mosca, R., Kamburov, A., Ghiassian, S. D., Yang, X., Ghamsari, L., Balcha, D., Begg, B. E., Braun, P., Brehme, M., Broly, M. P., ... Vidal, M. (2014). A proteome-scale map of the human interactome network. Cell, 159 (5), 1212-26. doi:10.1016/j.cell.2014.10.050  Peer Reviewed verified by ORBi Peer Reviewed verified by ORBi* These authors have contributed equally to this work. |
  | Blibek, K., Rambout, X., Beaufays, J., Lins, L., Dequiedt, F., & Twizere, J.-C. (2014). An interaction map for HTLV-1 Tax and PDZ-containing proteins. Retrovirology, 11, 101. doi:10.1186/1742-4690-11-S1-P101  Peer Reviewed verified by ORBi Peer Reviewed verified by ORBi |
  | Rambout, X. (2013). Construction of an interactomic map of Ets factors and identification of new functions in mRNA processing [Doctoral thesis, ULiège - Université de Liège]. ORBi-University of Liège. https://orbi.uliege.be/handle/2268/156708 |
  | Blibek, K., Rambout, X., beaufays, J., Lins, L., Dequiedt, F., & Twizere, J.-C. (29 June 2013). An interaction map for HTLV-1 Tax and PDZ-containing proteins [Poster presentation]. 16th International Conference on Human Retrovirology : HTLV and Related Viruses, Montréal, Québec., Canada. |
Rambout, X., Simonis, N., Brohée, S., Cherkaoui, M., Lebrun, M., Vidal, M., Kruys, V., Twizere, J.-C., Soin, R., & Dequiedt, F. (28 January 2013). Interactomic map of the Ets factors family : Identification of unexpected functions in mRNA processing [Poster presentation]. GIGA Day 2013. |
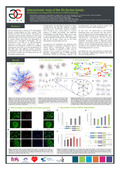  | Rambout, X., Simonis, N., Demoitié, P., Cherkaoui, M., Lebrun, M., Vidal, M., Kruys, V., Twizere, J.-C., & Dequiedt, F. (2012). Interactomic map of the Ets factors family : Identification of unexpected functions in mRNA processing. In Keystone symposium - Protein-RNA Interactions in Biology and Disease (C1).  Peer reviewed Peer reviewed |
  | Condé, C., Rambout, X., Lebrun, M., Lecat, A., Di Valentin, E., Dequiedt, F., Piette, J., Gloire, G., & Legrand, S. (2012). The inositol phosphatase SHIP-1 inhibits NOD2-induced NF-κB activation by disturbing the interaction of XIAP with RIP2. PLoS ONE. doi:10.1371/journal.pone.0041005  Peer Reviewed verified by ORBi Peer Reviewed verified by ORBi |
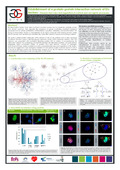  | Rambout, X., Simonis, N., Demoitié, P., Vidal, M., Twizere, J.-C., & Dequiedt, F. (29 April 2011). Establishment of an interactomic map of the Ets factors family: Towards a better understanding of their roles in oncogenic processes [Poster presentation]. IAP Network 28 : Signal integration mechanisms in health and disease, Bruxelles, Belgium. |
  | Rambout, X., Twizere, J.-C., & Dequiedt, F. (2010). Establishment of an interactomic map of the Ets factors family: Towards a better understanding of their roles in oncogenic processes. In Inserm Workshop: Interactomics: at the crossroads of biology and bioinformatics.  Peer reviewed Peer reviewed |
Lecat, A., Di Valentin, E., Fillet, M., Rambout, X., Twizere, J.-C., Dequiedt, F., Piette, J., & Legrand-Poels, S. (28 January 2010). NOD2 interactome [Poster presentation]. inflammation 2010, Luxembourg, Luxembourg. |
Rambout, X. (2008). Investigating Prostaglandin E2 activation of the p38 pathway of oesophageal squamous cell carcinoma [Master’s dissertation, ULiège. GxABT - Liège Université. Gembloux Agro-Bio Tech]. ORBi-University of Liège. https://orbi.uliege.be/handle/2268/155142 |